|
3D macromolecule structures are always rich sources of information, and we're well-equipped to help you extract meaning from any structure you've determined. We can help you with: searching for existing structural homologs; identification and characterization of protein:protein and protein:ligand interfaces; designing mutants to disrupt those interfaces; identification of conserved surface features as potential partner-interaction sites.
By way of example, this is the potassium-binding site of S.cerevisiae Sec12:
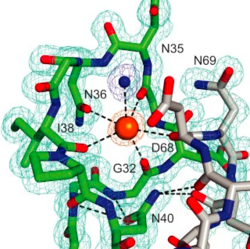
We discovered this metal-binding site while building the structure and subsequent crystallographic, structural and biochemical analysis confirmed it as potassium. Sec12's guanine exchange activity was found to be strongly modulated by potassium concentration, a previously unappreciated feature. See the paper in JBC.
Structure Data Mining
We have been developing a database-backed approach to making the most of the enormous amount of structural data available in the PDB: 95,000+ structures and increasing at a staggering rate that defies any individual's attempt to comprehend all of it.We have databases that summarize the biological context of structures from the deposited data itself (PDB), provides links between structure and function via Pfam and Gene Ontology databases, provides mapping between similar structures (DALI, CATH and SCOP databases) and are always in a strong position to leverage our own programs and experience to address pretty much any structure-based question that's on your mind. We offer custom-application programming for those questions if our ever-expanding database can't answer it immediately.